Modeling the Tumor Microenvironment with Graph Concept Learners
Using concept and geometric-deep learning to enable interpretable predictions of tumor tissue images.
Abstract
Heterogeneity is an emergent property of tumors, linked to cancer resistance and poor treatment outcomes (Marusyk et al., 2012). Geometric deep learning using graph representations has emerged as a promising approach to investigate tumor heterogeneity. Still, these approaches suffer from interpretability and transferability limitations (Rudin, 2019).
In this work, we propose a geometric deep-learning model with an enhanced interpretability framework to predict metadata from spatial datasets. Our approach draws inspiration from concept learning in computer vision and introduces concept graphs as a way of defining high-level postulates relevant to the task at hand (Cao et al., 2021).
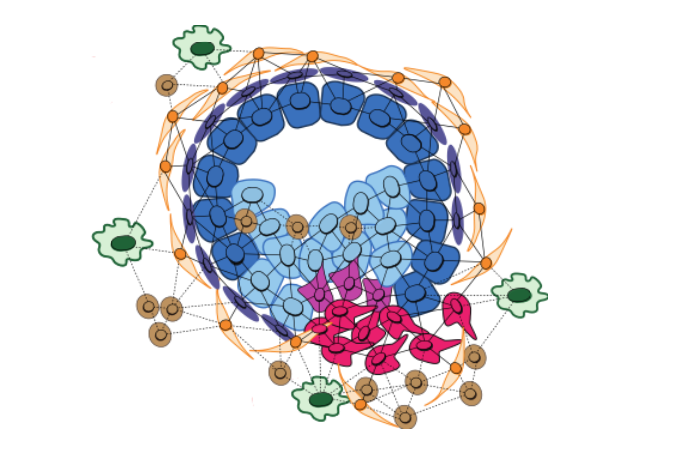
Cell graphs are a graphical representation of a tissue, where nodes represent cells and edges between nodes denote adjacency between cells in the high-dimensional omic space. Concept graphs are cell graphs constructed on a subset of all cells, where the filtering that induces a subset is defined by an underlying postulate. We propose to model the tumor microenvironment by constructing concept graphs that represent relevant postulates such as well-established cancer hallmarks (e.g., tumor-promoting inflammation) by constructing subgraphs including only cells that participate in the relevant interactions (e.g., tumor-immune cells).
Altogether, our approach decomposes each tumor sample into concept graphs and computes embeddings for each of them using a concept-specific graph neural network, and finally, integrates all embeddings in a downstream prediction engine with an attention mechanism (Vaswani et al., 2017). This mechanism highlights the relevance of each concept for a given prediction, thereby opening an avenue for interpretation.
The novelty of our approach lies in disentangling complex tumor graphs into interpretable concept graphs, allowing us to explore the relevance of inputs in terms of high-level concepts instead of low-level features like pixels or nodes in a graph. We applied our framework on a publicly available breast cancer imaging mass cytometry dataset (Jackson et al., 2020), achieving highly accurate and interpretable predictions of several clinically relevant metadata.
With spatial omics and multiplexed imaging data becoming increasingly popular, we hope that our approach will offer an interpretable and transferable framework that can assist in identifying cancer-related mechanisms and their relevance in clinical predictions.
Want to know more?
- 📖 Thesis (will be published soon).
- 📝 Paper (coming soon!).
- 🚀 Checkout the Github repo.
References
2021
2020
- Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancerNature Cancer, Feb 2020Number: 2 Publisher: Nature Publishing Group
2019
- Stop explaining black box machine learning models for high stakes decisions and use interpretable models insteadNature Machine Intelligence, May 2019Number: 5 Publisher: Nature Publishing Group
2017
2012
- Intra-tumour heterogeneity: a looking glass for cancer?Nature Reviews Cancer, May 2012Number: 5 Publisher: Nature Publishing Group