Abstract
Structural variants (SVs) are a very pervasive type of mutations present across cancers. Plenty of research has been done exploring how these aberrations affect differential expression and genomic architecture. However little research has been done concerning the overall effect of this type of mutations in the transcriptional regulatory programs. Building on the work of García-Cortés et al. (2018) and Espinal-Enríquez et al. (2017), we ask whether genes that are significantly copy number mutated are enriched in subtype specific breast cancer mutual information transcriptional networks.
We use the TCGA breast cancer data to build the networks with ARACNE and to determine which genes are significantly copy number mutated in each subtype using GISTIC2.0 (Mermel et al. 2011; Margolin et al. 2006). Then we perform two sided fisher tests enrichment analysis to determine whether these genes are over or under represented in our cancer transcriptional networks. Furthermore, we explore the question of whether these types of mutations could be shaping the observed network architecture. This question however is discussed but not addressed formally.
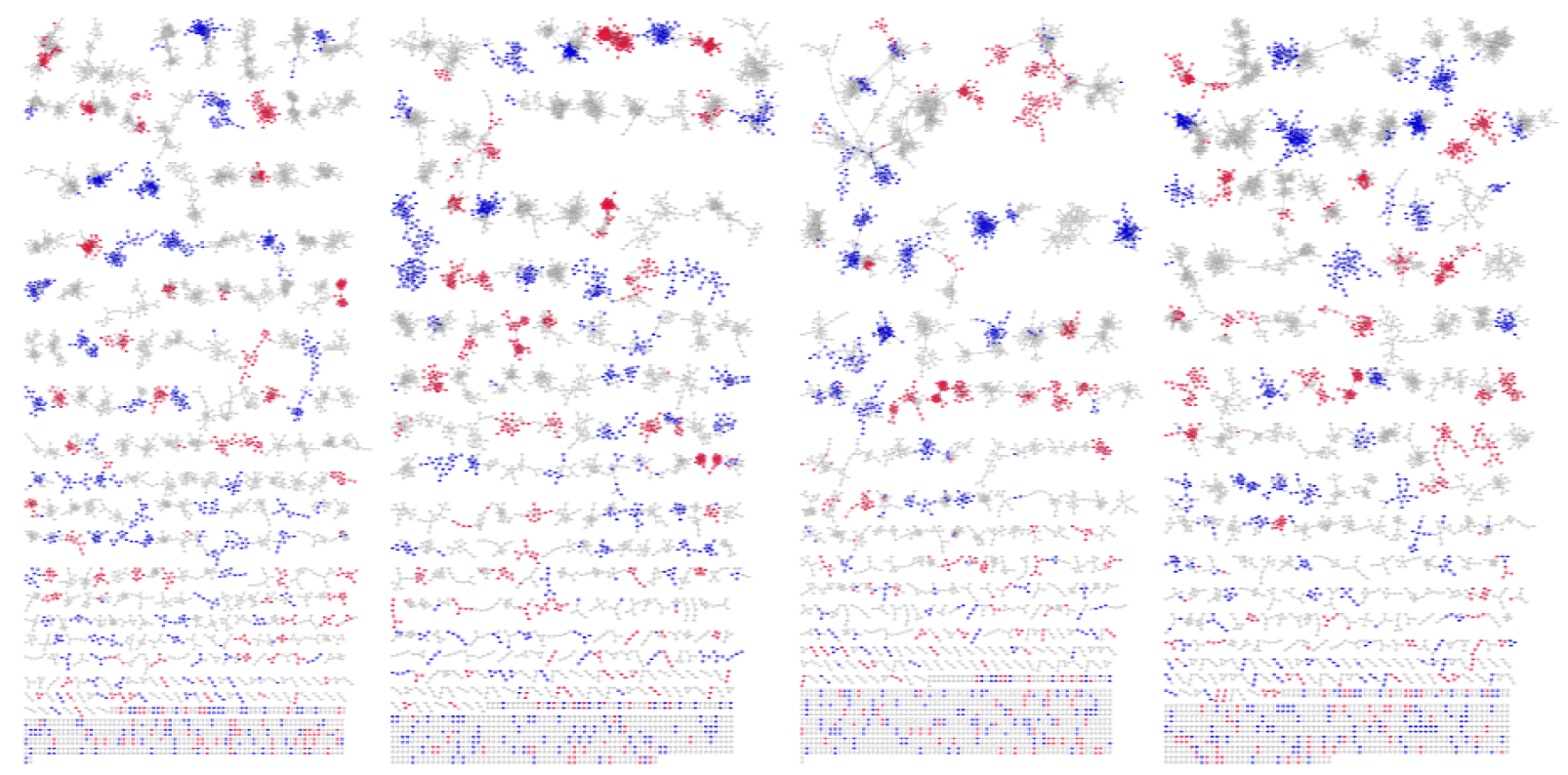
Each column represents a transcriptional network (13k most significant interactions) for each breast cancer molecular subtype (from left to right networks belong to Basal, Her2, LumA and LumB). Nodes dyed blue if said gene were found to be significantly deleted and red if significantly amplified.
Want to know more?
- 🚀 Check out the full report here.
References
2020
-
Gene co-expression is distance-dependent in breast cancer
Diana García-Cortés, Guillermo de Anda-Jáuregui, Cristóbal Fresno, and 2 more authors
Apr 2020
Pages: 399253 Section: New Results
Breast carcinomas are characterized by anomalous gene regulatory programs. As is well known, gene expression programs are able to shape phenotypes. Hence, the understanding of gene co-expression may shed light on the underlying mechanisms behind the transcriptional regulatory programs affecting tumor development and evolution. For instance, in breast cancer, there is a clear loss of inter-chromosomal (trans-) co-expression, compared with healthy tissue. At the same time cis- (intra-chromosomal) interactions are favored in breast tumors. In order to have a deeper understanding of regulatory phenomena in cancer, here, we constructed Gene Co-expression Networks by using 848 RNA-seq whole-genome samples corresponding to the four breast cancer molecular subtypes, as well as healthy tissue. We quantify the cis-/trans- co-expression imbalance in all phenotypes. Additionally, we measured the association between co-expression and physical distance between genes, and characterized the proportion of intra/inter-cytoband interactions per phenotype. We confirmed loss of trans- co-expression in all molecular subtypes. We also observed that gene cisco-expression decays abruptly with distance in all tumors in contrast with healthy tissue. We observed co-expressed gene hotspots, that tend to be connected at cytoband regions, and coincide accurately with already known copy number altered regions, such as Chr17q12, or Chr8q24.3 for all subtypes. Our methodology recovered different alterations already reported for specific breast cancer subtypes, showing how co-expression network approaches might help to capture distinct events that modify the cell regulatory program.
2017
-
RNA-Seq based genome-wide analysis reveals loss of inter-chromosomal regulation in breast cancer
Jesús Espinal-Enríquez, Cristóbal Fresno, Guillermo Anda-Jáuregui, and 1 more author
Scientific Reports, May 2017
Number: 1 Publisher: Nature Publishing Group
Breast cancer is a complex heterogeneous disease. Common hallmark features of cancer can be found. Their origin may be traced back to their intricate relationships governing regulatory programs during the development of this disease. To unveil distinctive features of the transcriptional regulation program in breast cancer, a pipeline for RNA-seq analysis in 780 breast cancer and 101 healthy breast samples, at gene expression and network level, was implemented. Inter-chromosomal relationships between genes resulted strikingly scarce in a cancer network, in comparison to its healthy counterpart. We suggest that inter-chromosomal regulation loss may be a novel feature in breast cancer. Additional evidence was obtained by independent validation in microarray and Hi-C data as well as supplementary computational analyses. Functional analysis showed upregulation in processes related to cell cycle and division; while migration, adhesion and cell-to-cell communication, were downregulated. Both the BRCA1 DNA repairing signalling and the Estrogen-mediated G1/S phase entry pathways were found upregulated. In addition, a synergistic underexpression of the γ-protocadherin complex, located at Chr5q31 is also shown. This region has previously been reported to be hypermethylated in breast cancer. These findings altogether provide further evidence for the central role of transcriptional regulatory programs in shaping malignant phenotypes.
2011
-
GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers
Craig H. Mermel, Steven E. Schumacher, Barbara Hill, and 3 more authors
Genome Biology, Apr 2011
We describe methods with enhanced power and specificity to identify genes targeted by somatic copy-number alterations (SCNAs) that drive cancer growth. By separating SCNA profiles into underlying arm-level and focal alterations, we improve the estimation of background rates for each category. We additionally describe a probabilistic method for defining the boundaries of selected-for SCNA regions with user-defined confidence. Here we detail this revised computational approach, GISTIC2.0, and validate its performance in real and simulated datasets.
2006
-
ARACNE: An Algorithm for the Reconstruction of Gene Regulatory Networks in a Mammalian Cellular Context
Adam A. Margolin, Ilya Nemenman, Katia Basso, and 4 more authors
BMC Bioinformatics, Mar 2006
Elucidating gene regulatory networks is crucial for understanding normal cell physiology and complex pathologic phenotypes. Existing computational methods for the genome-wide "reverse engineering" of such networks have been successful only for lower eukaryotes with simple genomes. Here we present ARACNE, a novel algorithm, using microarray expression profiles, specifically designed to scale up to the complexity of regulatory networks in mammalian cells, yet general enough to address a wider range of network deconvolution problems. This method uses an information theoretic approach to eliminate the majority of indirect interactions inferred by co-expression methods.