Microbiome-Analysis Software Tools for Qiime2
Microbiome-analysis software tool development with reproducibility at its core.
What I do
Since August 2023 I am working at the Food Systems Biotechnology group as a software engineer. My work in this group consist of developing and integrating bioinfomratics tools for metagenome-analysis with Qiime2.
Qiime2 is potent microbiome analysis tool that prioritizes transparency and reproducibility. It does so by featuring automated data tracking, a semantic type system, and a plugin system for expanded functionality.
My responsibilities in the devolving team include:
- 🔧 adding new functionalities to already existing plugins
- 👀 reviewing issues and PRs
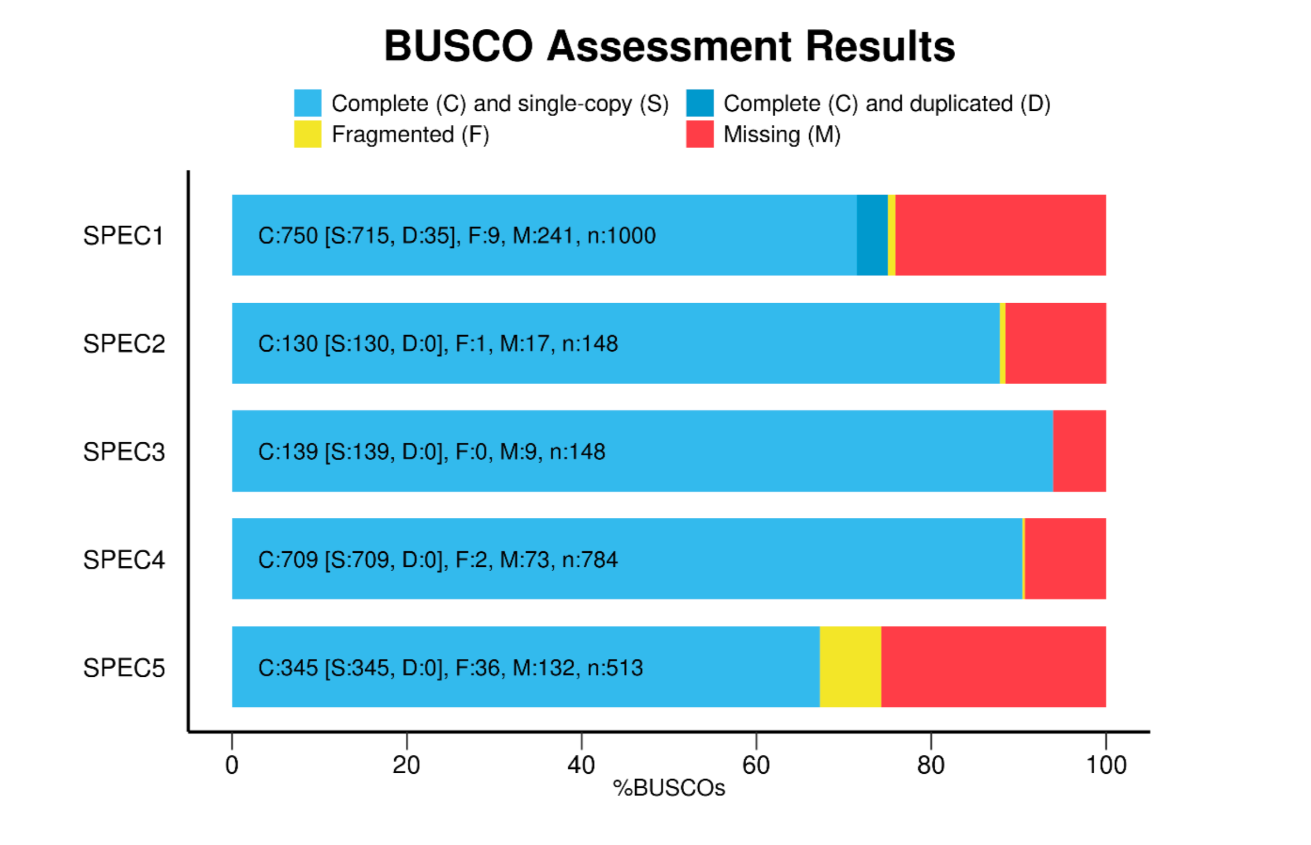
For example I recently added BUSCO to the q2-moshpit plugin (Manni, 2021). BUSCO is a tool to quantify the quality of assembled metagenomes based on the presence of universal single-copy orthologues. The goal was to build a wrapper for the existing tool to make it available within Qiime2. Furthermore we sought to present the results through costume interactive visualizations like this one.
Want to learn more?
- 🚀 Check out the Food Systems Biotechnology GitHub organization.
References
2021
- BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral GenomesMolecular Biology and Evolution, Jul 2021